Migration Genomics
Migration Genomics
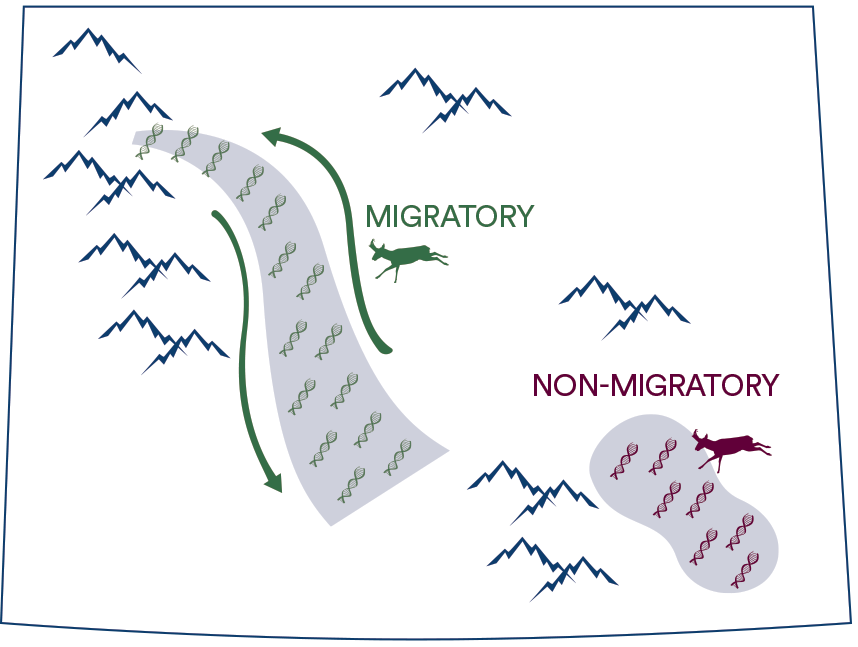
We use population genomics to investigate genetic differences among individuals in a population that have different seasonal migration strategies (i.e., migrants and non-migrants). Previous research has shown genetic segregation between migrants and non-migrants in the Yellowstone pronghorn population (Barnowe-Meyer et al. 2013).
Migration Basics
Animals make seasonal movements to find better food resources or to avoid harsh weather in a process we call migration. Seasonal migration is extremely variable across species – some animals travel very long distances, while others travel shorter distance; and some animals travel the exact same path to the same location every year, while others move opportunistically. There can even be variation within a species. For example, many species that migrate seasonally can be classified as partially migratory, meaning that within a population, some individuals make seasonal movements, while other individuals stay in the same area year-round.
What controls seasonal migration?
This question has yet to be answered definitively in the scientific literature, but we know the answer varies across species and is likely a combination of different factors. Migration can be controlled by:
- Genetics: some migratory traits are genetically programmed
- Example: European warblers have a genetic basis for migratory restlessness, or the urge to migrate (Berthold and Querner 1981)
- Social learning: some animals learn to migrate from their relatives
- Example: White-tailed deer typically utilize the same migratory routes and seasonal ranges as their mothers (Nelson 1998)
- Environmental cues: some animals use environmental cues to determine when or where to migrate seasonally
- Example: Barnacle Geese “surf the green wave”, meaning that during spring migration they time their travel so that they arrive at each stopover site when forage quality is highest (Van der Graaf et al 2006)
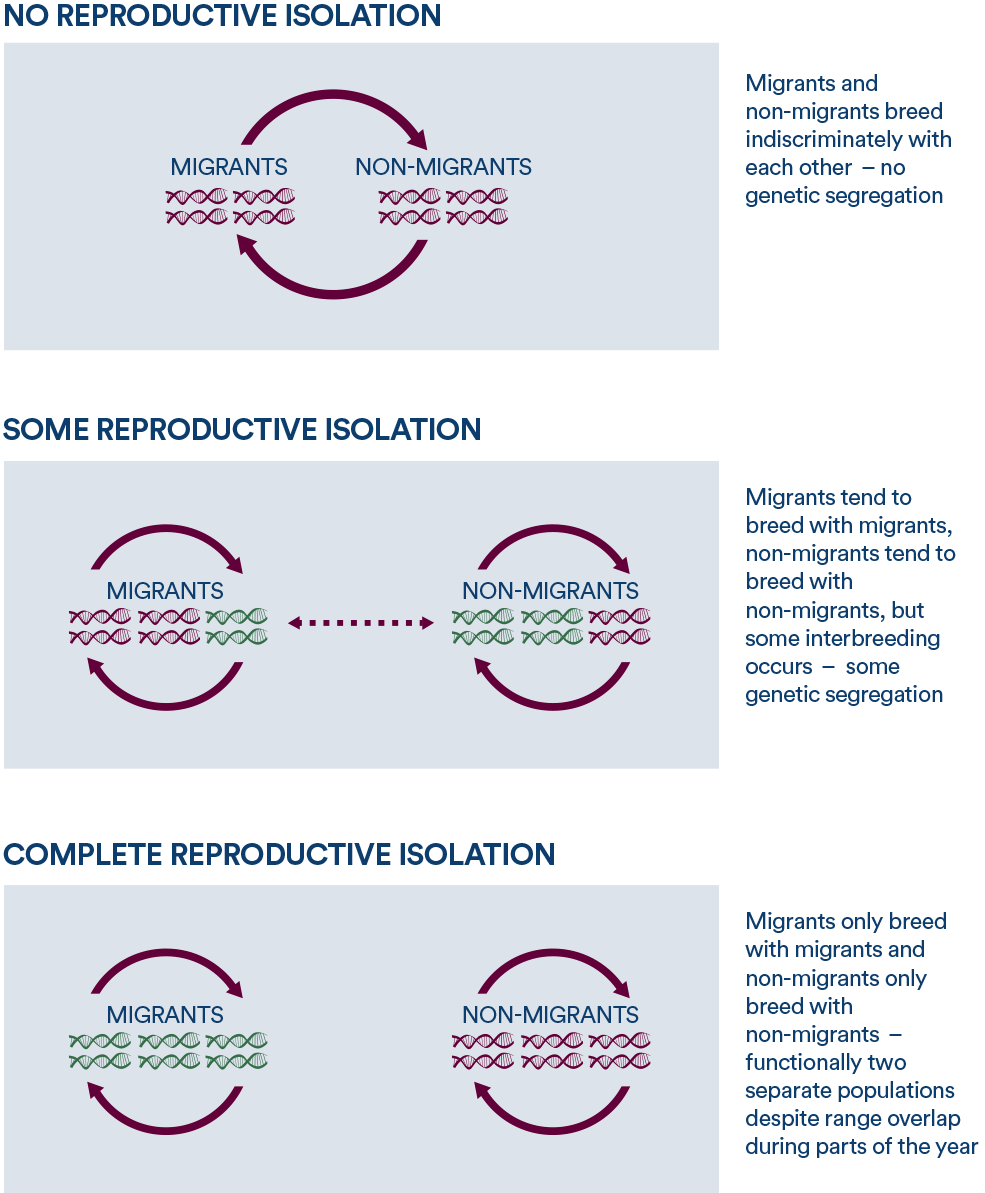
These three generalized scenarios represent gene flow (interbreeding, genetic interchange) caused by different levels of reproductive isolation among migration strategies.
Ungulate Migration & Population Genomics
In our lab, we use population genetics to study ungulate migration. After determining whether individual animals are migratory or non-migratory using GPS tracking data, we can investigate genetic differences between these two groups. This work requires collaboration with other researchers who study seasonal migration to help us determine which individuals migrate and which do not. To learn more about our pronghorn migration genomics project, see our pronghorn research page.
References
Berthold, P. and U. Querner. 1981. Genetic basis of migratory behavior in European warblers. Science 212:77-79. DOI: 10.1126/science.212.4490.77.
Nelson, M. E. 1998. Development of migratory behavior in northern white-tailed deer. Canadian Journal of Zoology, 76:426-432.
Van der Graaf, A. J., Stahl, J., Klimkowska, A., Bakker, J. P., and R. H. Drent. 2006. Surfing on a green wave-how plant growth drives spring migration in the Barnacle Goose Branta leucopsis. ARDEA-WAGENINGEN 94:567.
Barnowe-Meyer, K. K., White, P. J., Waits, L. P., & Byers, J. A. (2013). Social and genetic structure associated with migration in pronghorn. Biological Conservation, 168, 108–115. http://doi.org/10.1016/j.biocon.2013.09.022

